In a recent study published in the journal npj Vaccines, researchers investigated whether the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) messenger ribonucleic acid (mRNA) vaccine could alter the breast milk microbiome and ways in which the changes impact breast milk antibody titers.
Vaccine-induced antibodies in breast milk confer immunity to newborns against SARS-CoV-2. However, concerns have been raised about nursing women’s possible short- and long-term adverse effects and vaccine-elicited alterations in the makeup of the human milk microbiota, which helps mold the initial-life microbiota.
 Study: COVID-19 mRNA vaccine-mediated antibodies in human breast milk and their association with breast milk microbiota composition. Image Credit: PenguinDaddy / Shutterstock
Study: COVID-19 mRNA vaccine-mediated antibodies in human breast milk and their association with breast milk microbiota composition. Image Credit: PenguinDaddy / Shutterstock
About the study
In the present study, researchers evaluated the association between BNT162b2 vaccine-induced antibodies and the microbiome in human milk.
The study included 49 breastfeeding mothers who received two BNT162b2 vaccinations in Hong Kong from June to August 2021 and were serologically negative for SARS-CoV-2 before vaccination. Samples of human milk were self-obtained by the participants before vaccination, a week after the initial vaccination, a week after the subsequent vaccination, and a month after the subsequent vaccination.
For microbial analysis, deoxyribonucleic acid (DNA) was obtained from the human milk samples. Amplicon sequence variants (ASVs) were created using the QIIME2 pipeline. Enzyme-linked immunosorbent assays (ELISA) were performed to measure SARS-CoV-2 spike (S) glycoprotein-targeted immunoglobulin G (IgG) and IgA titers in human milk and assess immunological responses to the BNT162b2 vaccine at different phases of the vaccination regimen.
Further, 16 S ribosomal ribonucleic acid (rRNA) amplicon sequencing was performed to investigate whether the microbial community in human milk was related to the anti-S receptor-binding domain (RBD) antibody titers in human milk evoked by the BNT162b2 vaccination. The analysis removed samples that lacked sequencing information. Linear mixed models were used to examine and compare the composition and diversity of the microbiome in human milk over different periods of time.
Furthermore, linear discriminant analysis (LEfSe) was performed to identify bacterial species that were abundant at the post-vaccination time points. The researchers also investigated whether there were taxonomic changes in the baseline microbiome of those with low and high IgA titers. The team determined the relationship between anticipated microbial functional pathways and anti-SARS-CoV-2 IgA titers a week after the second vaccination to study the potential mechanisms through which the pre-vaccination human milk microbiota impacts antibody titers.
The team constructed classification models to predict levels of anti-SARS-CoV-2 antibodies in human milk a week after the second vaccination using baseline human milk microbiome abundances based on significant differential microbial predictors and combining the microbial estimators with demographics (age, dosing interval, and intramuscular analgesia usage at delivery).
Results
In total, 175 breast milk samples provided by 44 women were examined. The median age of the participants was 36 years, and the median breastfeeding stage at which the milk samples were obtained was 37 weeks. Among the participants, 44% had cesarean section deliveries. The median time between vaccines was 21 days.
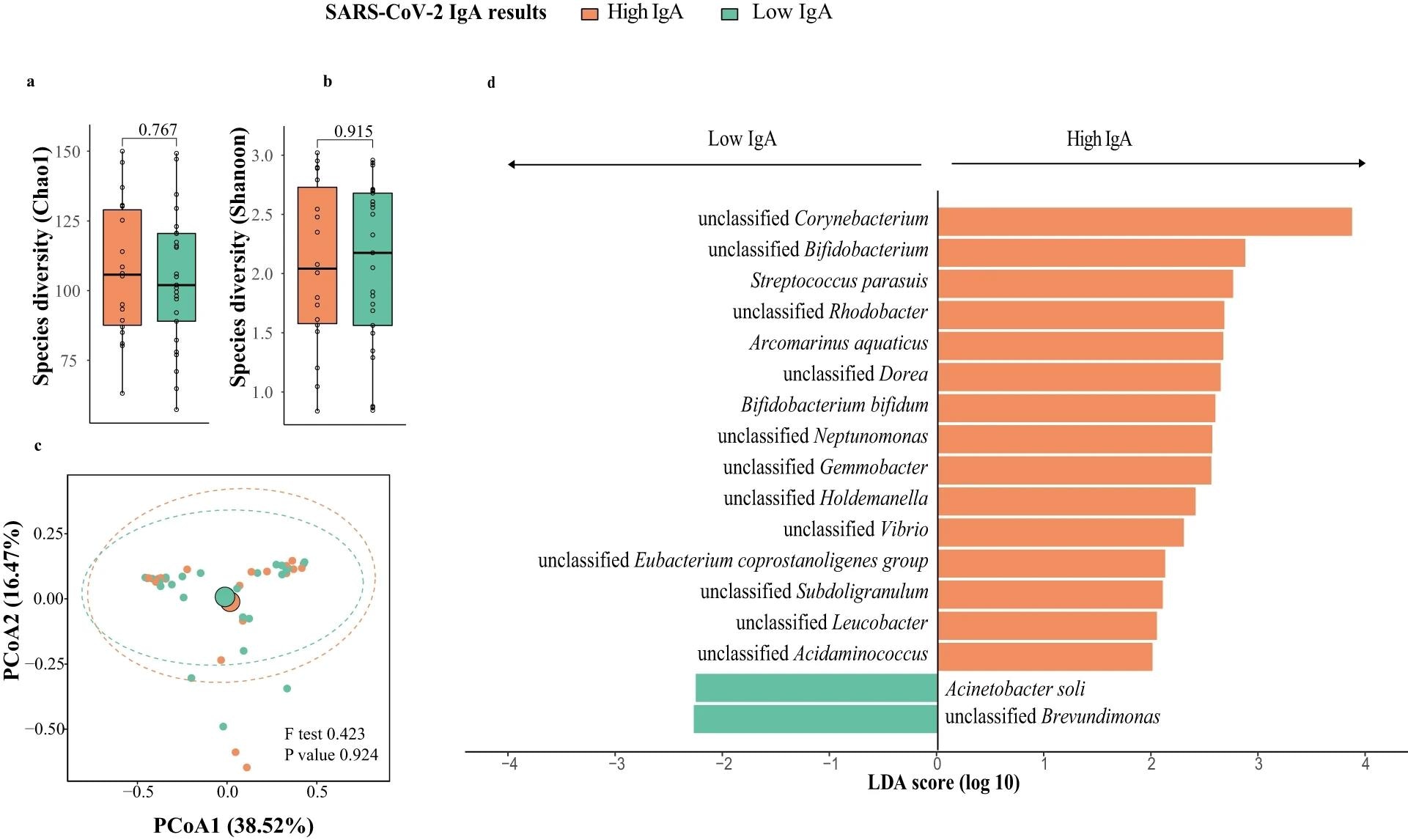 Breast milk microbiota composition in mothers with high and low responses at one week post-second dose of BNT162b2 (N = 43; High IgA: 18, Low IgA: 25). a Bacteria diversity. b Bacteria richness. P value comparing the diversity and richness were given by Wilcoxon rank-sum test. c Principal coordinates analysis (PCoA) of breast milk microbiota composition of mothers with high- and low-IgA levels at one week post-second dose of BNT162b2. p value was given by PERMANOVA. d Linear discriminant analysis effect size analysis of discriminant taxa in breast milk microbiome of mothers with high- and low-IgA levels at one week post-second dose of BNT162b2. LDA linear discriminant analysis. Elements on boxplots: centre line, median; box limits, upper and lower quartiles; whiskers, 1.5×IQR.
Breast milk microbiota composition in mothers with high and low responses at one week post-second dose of BNT162b2 (N = 43; High IgA: 18, Low IgA: 25). a Bacteria diversity. b Bacteria richness. P value comparing the diversity and richness were given by Wilcoxon rank-sum test. c Principal coordinates analysis (PCoA) of breast milk microbiota composition of mothers with high- and low-IgA levels at one week post-second dose of BNT162b2. p value was given by PERMANOVA. d Linear discriminant analysis effect size analysis of discriminant taxa in breast milk microbiome of mothers with high- and low-IgA levels at one week post-second dose of BNT162b2. LDA linear discriminant analysis. Elements on boxplots: centre line, median; box limits, upper and lower quartiles; whiskers, 1.5×IQR.
Anti-S protein IgG and IgA levels in human milk were unchanged after the initial dosage in comparison to pre-vaccination levels. A week after the second vaccination, the titers were raised [median interquartile range (IQR), IgA optical density (OD) values: 0.1 versus 0.2; IgG antibody OD values: 0.1 compared to 0.2]. Although IgA titers returned to baseline levels after a month of the second vaccination, IgG titers were considerably higher (IgG antibody OD values: 0.1 versus 0.2).
An area under the receiver operating characteristic curve (AUC) value of 0.7 was obtained to estimate anti-SARS-CoV-2 S IgA titers a week after the second vaccination by assessing the baseline human milk microbial composition. Furthermore, a moderately strong association was observed between the two antibodies in human milk. Human milk composition and richness were dynamically altered during the vaccination program; however, Bifidobacterium beneficial bacterial abundance remained unaltered post-vaccination.
Chao1 bacteria richness increased, but not Shannon diversity, a week after the first vaccination. However, a week following the second vaccination, microbial richness decreased to its initial levels. At study initiation, Shannon richness among individuals with high IgA titers was significantly greater compared to that among their low-IgA counterparts. Proteobacteria, Firmicutes, and Actinobacteria were highly prevalent phyla. Firmicutes were more abundant in low-IgA individuals, but proteobacteria counts were lower.
The researchers discovered 23 species that differed in abundance between high- and low-IgA individuals, with Corynebacterium kroppenstedti and Neisseria having the most profound impact on enrichment. The stage of breastfeeding did not affect the relative quantity of these markers. Taxonomic alterations in the microbiome were also discovered a week after the second immunization. There were 437 functional microbial pathways suggested, with nine being substantially enriched in high-IgA people. Higher abundances of certain bacterial markers were linked to sugar metabolism pathways and super-pathways for arginine and polyamine production.
Overall, the study findings showed that the composition of the human milk microbiome and post-vaccination alterations were associated with BNT162b2-conferred humoral protection against SARS-CoV-2. The impact of the human milk microbiome on pathways associated with immunomodulatory metabolic activities, including the increased abundance of probiotic Bifidobacteria, may contribute to increased anti-SARS-CoV-2 antibody levels. These changes may provide additional immunological protection to infants through the vertical transmission of antibodies and beneficial microbiota.
Journal reference:
- Zhao, S., Lok, K.Y.W., Sin, Z.Y., et al. COVID-19 mRNA vaccine-mediated antibodies in human breast milk and their association with breast milk microbiota composition, npj Vaccines 8, 151 (2023) , DOI: https://doi.org/10.1038/s41541-023-00745-4, https://www.nature.com/articles/s41541-023-00745-4

 PARENTING TIPS
PARENTING TIPS PREGNANCY
PREGNANCY BABY CARE
BABY CARE TODDLERS
TODDLERS TEENS
TEENS HEALTH CARE
HEALTH CARE ACTIVITIES & CRAFTS
ACTIVITIES & CRAFTS

